Inter-session Alignment
Inter-Session Alignment and projection calculation
After motion correction we have to calculate relevant projections — projections of blood vessels and neurons used for alignment and downstream extraction — align the sessions, and then concatenate them. Alternatively, we can also process individual files independently:
To achieve this, CaliAli performs the following steps:
- Detrend each imaging session.
- Calculate projections of the blood vessels and neurons.
- Calculate displacement fields to align sessions using blood vessels and neurons.
- Evaluates blood vessel (BV) similarity and switches to neuron-based alignment if necessary.
- Apply displacement field to each video session.
- Standardized pixels and concatenate aligned videos:
This is done by executing;
Output File
Running this function produces a .mat file with the _Aligned tag, containing the aligned and concatenated video and the CaliAli_options structure. By default, the output file name matches the input video, and the save path defaults to the location of the input video. A typical file name may be 'Last_session_name"_ds_mc_Aligned.mat. This will also create one .mat file with the _det tag for each input session containing the detrended data and the relevant projections.
Even if we do not require to align sessions we still need to calculate relevant projections, as these frames are reused in later steps. This is done by executing:
Output File
Running this function produces a .mat file with the _det tag, containing the aligned and concatenated video and the CaliAli_options structure. By default, the output file name matches the input video, and the save path defaults to the location of the input video. A typical file name may be vid_01_ds_mc_det.mat.
Evaluating Alignment Performance:
Console outputs:
While running CaliAli_align_sessions(), the command window now uses coloured output (via cprintf) to highlight warnings and status information. A typical run looks like:
Blood-vessel similarity score: 5.376
Calculating correlation of the Neurons projections...
Processing: 100% |############| 6/6it [00:00:00<00:00:00, 32.27 it/s]
Correlation between Neurons projections is good!
Lowest spatial correlation: 0.491
-
Blood-vessel similarity score
-
Spatial correlation value
Should CaliAli detect per-session frame size changes (for example, if a user cropped one file manually), it now raises an informative message and falls back to safe handling rather than aborting.
Post alignment:
To visually confirm the alignment performance, load the *_Aligned.mat file in MATLAB:
First, load the alignment options from a saved .mat file.
For the demo file (assuming your MATLAB path includes the Demo folder) this would be:
'v4_mc_ds_Aligned.mat' with the path to your specific file.
You can evaluate the alignment performance by running the following lines:
% Get BV-score:
fprintf('BV Score: %.4f\n', CaliAli_options.inter_session_alignment.BV_score);
% Get alignment metrics: Mean Correlation score and Crispness (Higher the
% better)
Alignment_metrics = CaliAli_options.inter_session_alignment.alignment_metrics
plot_alignment_scores(CaliAli_options)
% View aligned projections:
P = CaliAli_options.inter_session_alignment.P; %extract the aligned projections
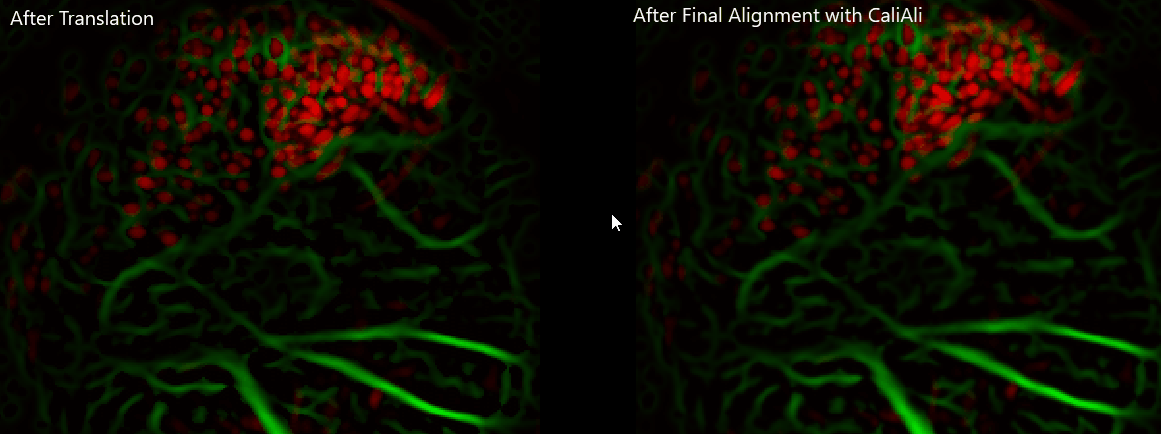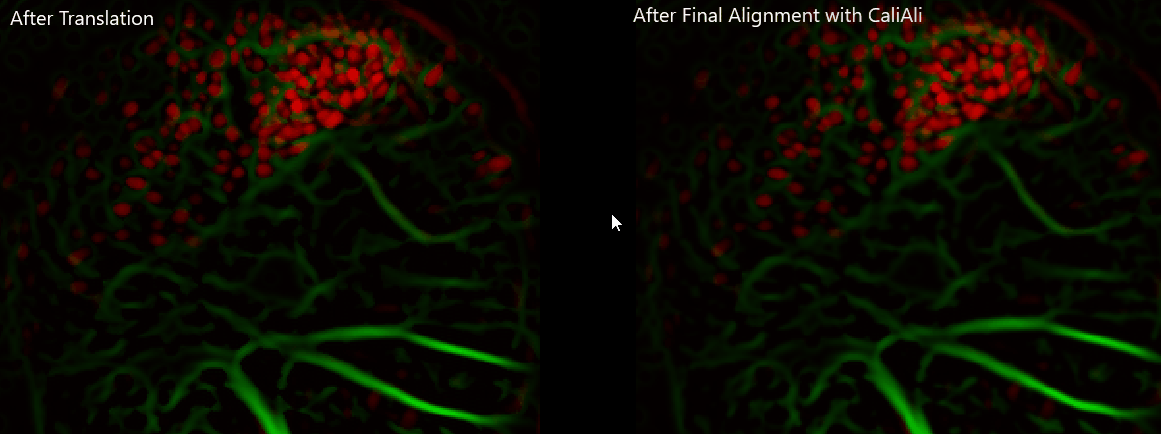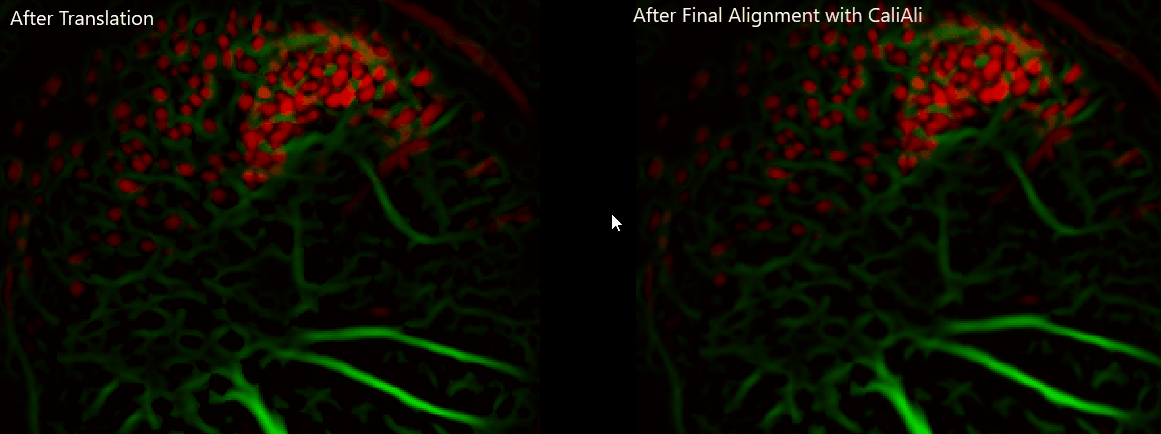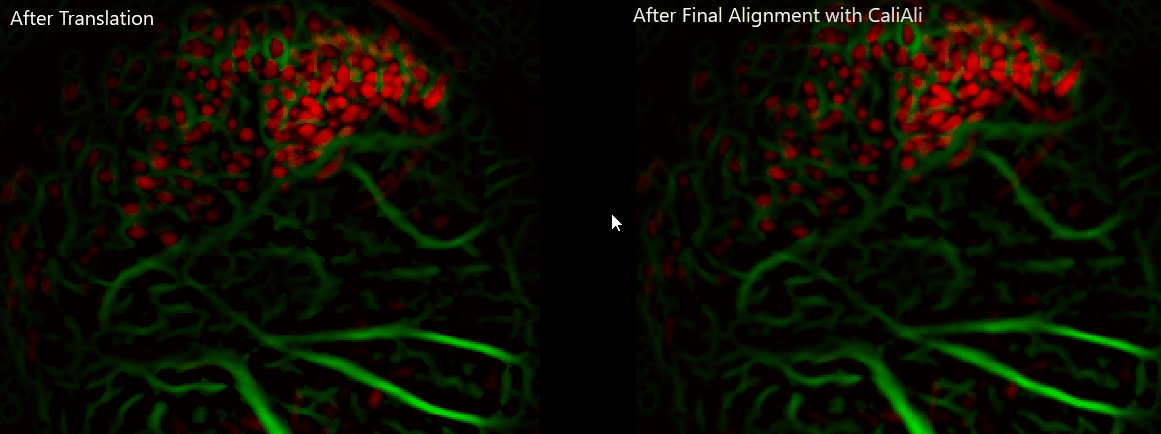
frame=plot_P(P); %Create video of the Aligned projections before and after CaliAli.
The most important components is the table P, which is structured as follow:
| Column | Description |
|---|---|
Original |
Projections before alignment |
Translation |
Projections after translation |
Multi-Scale |
Projections after multi-scale alignment |
Final |
Projections after final alignment |
Each column contains a nested table with projections organized as 3D or 4D arrays, representing data from each session:
| Column | Description |
|---|---|
Mean |
Mean frame of each session |
BloodVessels |
Blood vessels projection of each session |
Neurons |
Neuron projections of each session |
PNR |
PNR projections of each session |
BV+Neurons |
Blood vessels and neurons projection of each session |
You can visualize these projections with the following commands:
P = CaliAli_options.inter_session_alignment.P; %extract the aligned projections
frame=plot_P(P); %Create video of the Aligned projections before and after CaliAli.
Important
Please visually verify that sessions are correctly aligned. If you detect noticeable displacement in the field of view it means that CaliAli is not suitable for this data.
After finishing inter-session alignment or individual sessions processing you can proceed to Extract Calcium Traces with CaliAli